
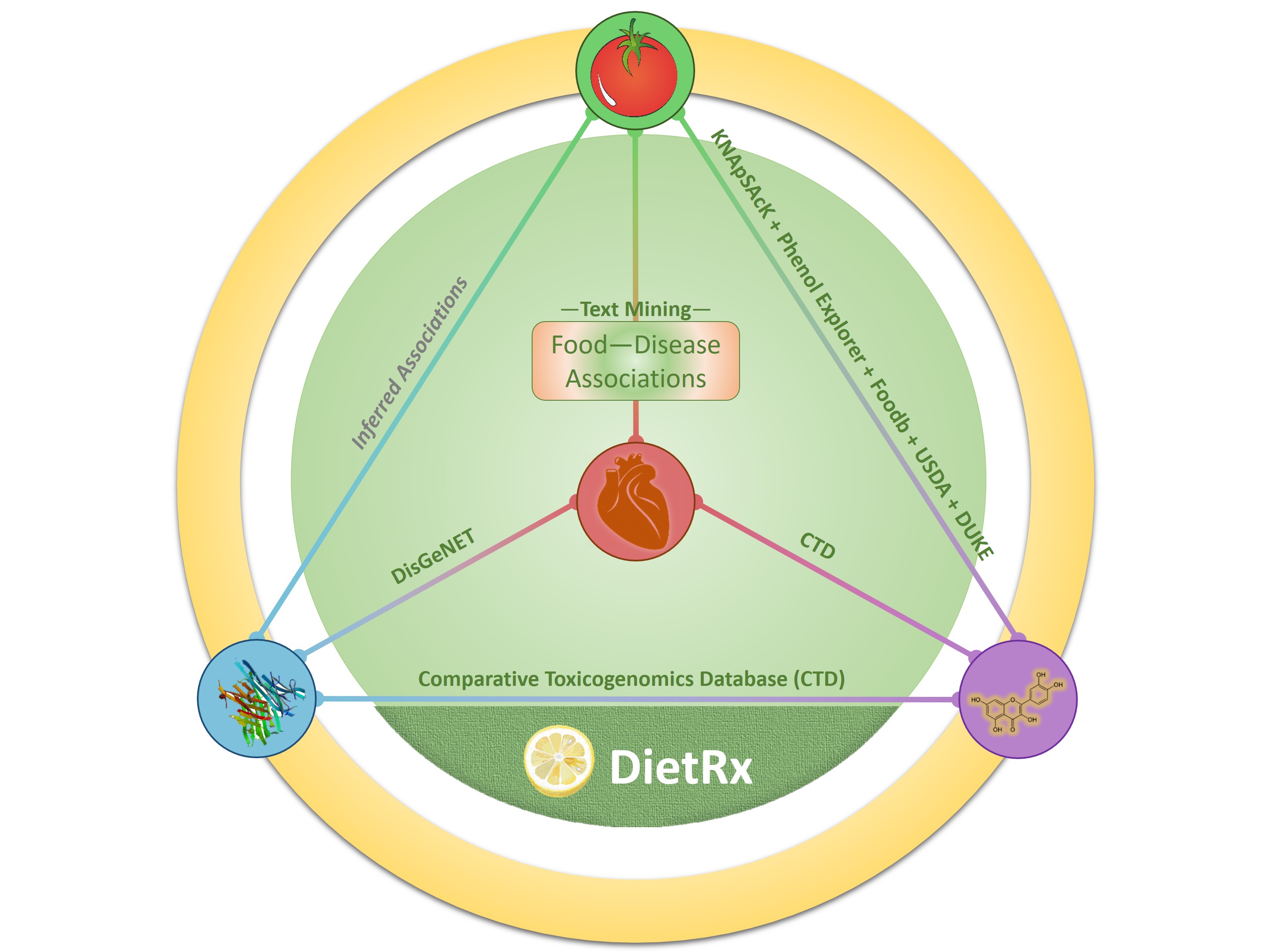
Explore Interrelationships Among Foods, Diseases, Genes and Chemicals

Explore Interrelationships Among Foods, Diseases, Genes and Chemicals
Contact us if you have more questions

| Name | Position | Affiliation | Contribution |
|---|---|---|---|
| Ganesh Bagler | Project Head | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Idea conception, Project design and management, Database design and implementation |
| Rudraksh Tuwani | Research Assistant | Center for Computational Biology, Indraprastha Institute of Information Technology | Data Curation, Database design, Development of DietRx Web Resource, Data Visualisation and Data Analytics |
| Neelansh Garg | Summer Research Intern | Center for Computational Biology, Indraprastha Institute of Information Technology | Database design, Development of DietRx Web Resource |
| Rakhi N K | PhD Research Scholar | Department of Bioscience and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur | Manual data compilation, Annotation and curation, Quality Check, Analysis |
| Rujhil Jain | M.Tech | Department of Computer Science And Engineering, IIIT-Delhi, Delhi | Contributed to backend development, database enhancements, and system optimization |
| Sneha Nandi | M.Tech | Department of Computer Science And Engineering, IIIT-Delhi, Delhi | Focused on data handling, backend consistency, and performance improvements |
| Shriyansh Bhardwaj | M.Tech | Department of Computer Science And Engineering, IIIT-Delhi, Delhi | Enhanced the user interface, optimized web functionalities, and supported system integration |