BitterSweet FAQs
Contact us if you have more questions
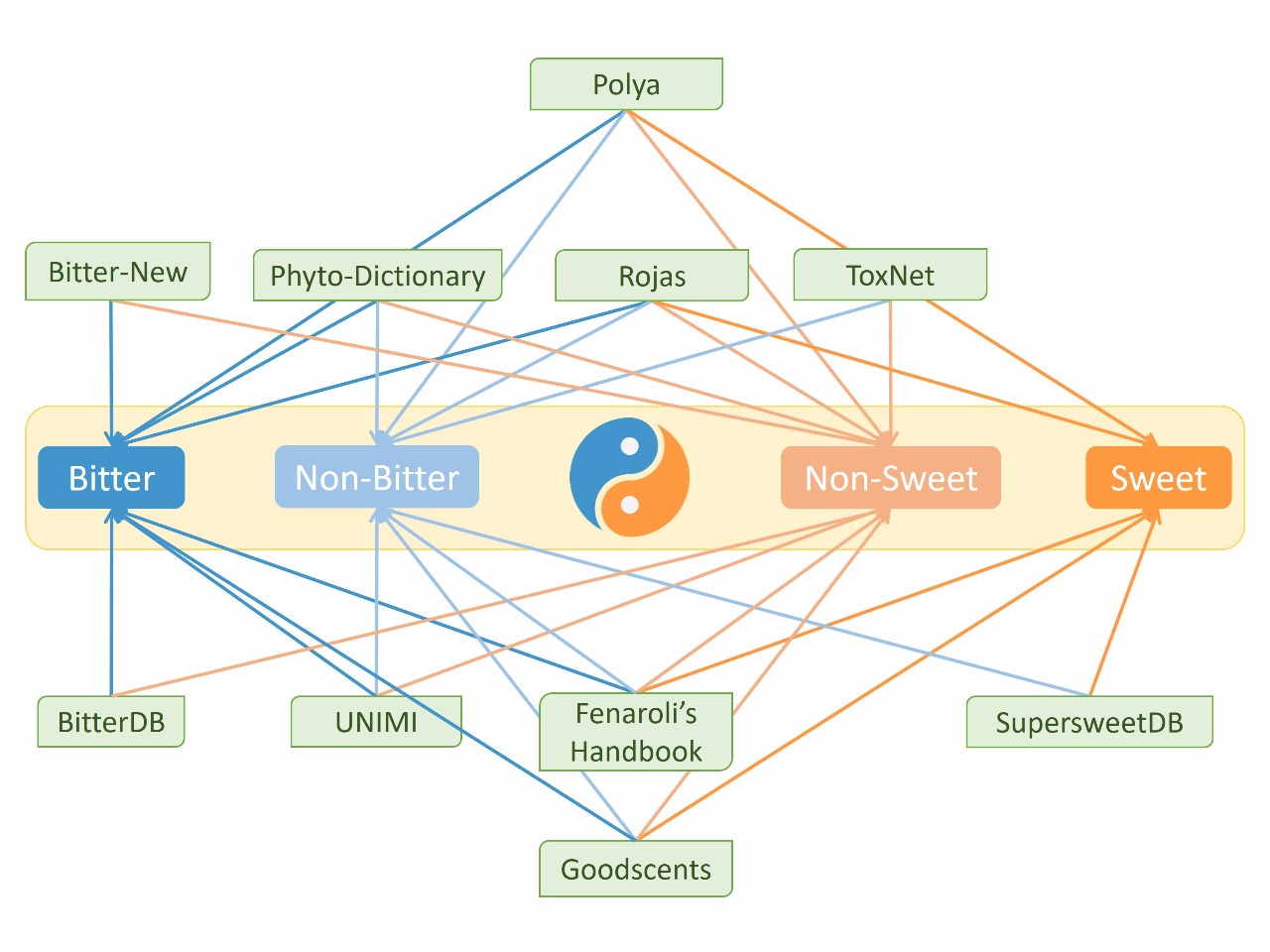
BitterSweet integrates information about a large number of bitter, sweet and tasteless molecules curated from a variety of different sources. These include but are not limited to commonly used databases such as BitterDB, SuperSweetDB, Toxnet and The Goodscents Company Database. In addition to these, there was also a careful curation carried out from other relevant sources like - Fenaroli's Handbook of Flavor Ingredients, Polya’s Biochemical Targets of Plant Bioactive Compounds and Phytochemical Dictionary.
Search of molecules is available by the use of the following identifiers -
- Common Name
- IUPAC Name
- PubChem ID
- Functional Group
- Structure Search (using JSME Molecular Editor)
An empty query in the BitterSweet search module will result in listing of all the molecules present in the entire database or a selected set of databases.
If you wish to check out the list of all possible molecules in BitterSweet, please Click Here
If you wish to check out the list of all possible molecules in BitterSweet, please Click Here
In addition to the regular search, advanced search is a more comprehensive search based on following molecular properties:
- Molecular Weight
- Hydrogen Bond Acceptor
- Hydrogen Bond Donor
- Number of Rings
- Rotatable Bond
- Number of Aromatic Bonds
- ALogP
ZINC is a free public resource for discovery of drug-like molecules. The database contains over twenty million commercially available molecules in biologically relevant representations that may be downloaded in popular ready-to-dock formats and subsets. The similarity search done against ZINC compares the query molecules with each of the molecules in the database. Depending upon the similarity (90% is the default threshold), it displays similar compounds. For further information on ZINC database please visit: ZINC Homepage
FlavorDB (Garg et al; FlavorDB: a database of flavor molecules, Nucleic Acids Research, January 2018) is a comprehensive resource of flavor molecules from natural sources consisting of over 25000 molecules of various tastes and odours. Search similar in FlavorDB identifies structurally similar molecules matching to the query molecules above the threshold similarity (90%). For further information on FlavorDB please visit: FlavorDB Homepage
Please refer to queries pertaining to ‘Serach Similar in ZINC’ and ‘Search Similar in FlavorDB’ as well as the How-To section.
The ‘functional group’ is an atom, or a group of atoms that has similar chemical properties whenever it occurs in different compounds. It defines the characteristic physical and chemical properties of families of organic compounds. (http://goldbook.iupac.org/F02555.html)
The functional groups were obtained using Checkmol, a free and an open source tool, checkmol, detect and assign the functional group information on any small molecules with 2D coordinates. The Checkmol is a command-line utility program, which reads molecular structure files in different formats and analyzes the input molecule for the presence of various functional groups.
Analysis of Functional Groups in Organic Molecules, http://merian.pch.univie.ac.at/~nhaider/cheminf/cmmm.html
N. Haider, “Functionality pattern matching as an efficient complementary structure/reaction search tool: an open-source approach”, Molecules 15 (8) (2010) 5079–5092. (http://www.mdpi.com/1420-3049/15/8/5079)
List of functional groups generated by Checkmol :- http://merian.pch.univie.ac.at/~nhaider/cheminf/fgtable.pdf
The “BitterSweet Predict” predicts whether the query molecules is Bitter, Sweet, Bitter-Sweet or ‘Nether Bitter or Sweet’. Starting with its SMILE/PubChem/ZINC the tool uses a state of the art model for predicting its “Bitter Probability” and “Bitter Probability” to further classify it into the following categories - Bitter, Sweet, Bitter-Sweet or Tasteless.
The following is a list of 10 sweet molecules of Mango from FlavorDB which you can yourself try out -
The following is a list of 10 sweet molecules of Mango from FlavorDB which you can yourself try out -
| SMILE ID | Name |
|---|---|
| CC1CCC(C(C1)O)C(C)C | Neomenthol |
| CC(=O)C(=O)C | 2,3-Butanedione |
| CCC1C(=O)C(=C(O1)C)O | 2-Ethyl-4-Hydroxy-5-Methyl-3(2H)-Furanone |
| CCCCCC(=O)C | 2-Heptanone |
| CCC(C)COC(=O)C | 2-Methylbutyl Acetate |
| CCCC(=O)C | 2-Pentanone |
| CCCC(=O)CC | 3-Hexanone |
| C1=CC=C(C=C1)CCC(=O)O | 3-Phenylpropanoic Acid |
| C(C1C(C(C(C(O1)OC2C(OC(C(C2O)O)O)CO)O)O)O)O | Alpha-Maltose |
| C1=CC=C(C=C1)CO | Benzyl Alcohol |
A reasonable selection of threshold for any binary classifier is set at 0.5. The False Positive Rate against different threshold values evaluated on the Test Set is specified below.
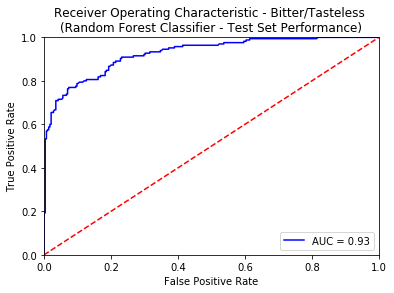
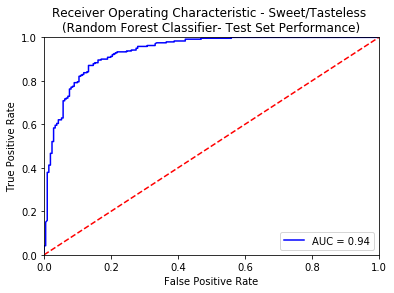


| False Positive Rate - Bitter | Threshold Value - Bitter | False Positive Rate - Sweet | Threshold Value - Sweet |
|---|---|---|---|
| 0 | 0.98 | 0 | 1 |
| 0 | 0.87 | 0 | 0.98 |
| 0 | 0.74 | 0.01 | 0.97 |
| 0.01 | 0.67 | 0.02 | 0.94 |
| 0.06 | 0.5 | 0.03 | 0.86 |
| 0.09 | 0.46 | 0.06 | 0.77 |
| 0.15 | 0.42 | 0.09 | 0.71 |
| 0.2 | 0.35 | 0.12 | 0.62 |
| 0.27 | 0.28 | 0.16 | 0.5 |
| 0.35 | 0.21 | 0.21 | 0.44 |
| 0.47 | 0.15 | 0.28 | 0.37 |
| 0.6 | 0.09 | 0.41 | 0.23 |
| 0.7 | 0.05 | 0.59 | 0.14 |
| 0.82 | 0.02 | 0.82 | 0.05 |
In addition to the standard tricks of avoiding overfitting (like Regularization, KFold Cross Validation), we also created an external validation set known as the Gold Standard from the molecules curated from 3 sources namely - Phyto Dictionary, Bitter New and UNIMI (These represent a general set of molecules which are known to have some sort of biological reactions with human body.). A high AUC-ROC value of 0.87 on these molecules ensured the robustness of our classification models.
Since we are using a machine learning model in the backend to generate predictions for a given molecule or a set of molecules, it is to be noted that the quality of these predictions is limited by the molecular properties and their relatibility to those of which were used to train these models. For instance, if the database is limited to the molecules whose molecular weight is less than 1000, then the prediction for an unknown molecule whose molecular weight far exceeds that value might not be reliable.
A: JMol is an open-source Java viewer for 3D chemical structures. It is a free and open source software and runs on Windows, Mac OS X, Linux and Unix systems. Some of the features Jmol include 3D visualisation, provision for zooming, and MOL2 download. For further information please visit the Jmol website.
A modern browser with JavaScript enabled.
A modern web browser with JavaScript enabled.
We are using cookies to provide statistics that help us give best experience for our site.
Yes, it has been tested to run successfully on Android as well as iOS platform.

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
| Name | Position | Affiliation | Contribution |
|---|---|---|---|
| Ganesh Bagler | Project Head | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Idea conception, Project design and management, Database design and implementation |
| Rudraksh Tuwani | Research Assistant | Center for Computational Biology, Indraprastha Institute of Information Technology | Text Mining, Database design, Development of BitterSweet Web Resource, Data Visualisation and Data Analytics |
| Somin Wadhwa | Research Intern | Maharaja Agrasen Institute of Technology, Guru Gobind Singh Indraprastha University, New Delhi | Predictive Modelling, Data Analytics, Development of BitterSweet Web Resource. |
| Nishant Kumar Singh | Research Intern | Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Redevelopment of the BitterSweet website. |
| Vaishvi Verma | Research Intern | Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Redevelopment of the BitterSweet website. |
Rudraksh Tuwani, Somin Wadhwa and Ganesh Bagler*, BitterSweet: A database of curated and predicted bitter and sweet molecules (2018),
http://cosylab.iiitd.edu.in/bittersweet
*Corresponding Author
http://cosylab.iiitd.edu.in/bittersweet
*Corresponding Author