Frequently Asked Questions (FAQs)
Q1: What are the various ingredient categories within FlavorDB?
- Additive
- Animal Product
- Bakery
- Beverage
- Beverage Alcoholic
- Beverage Caffeinated
- Cereal
- Maize
- Dairy
- Dish
- Essential Oil
- Fish
- Seafood
- Flower
- Fruit
- Berry
- Fruit Citrus
- Fruit Essence
- Fungus
- Herb
- Meat
- Legume
- Nut
- Seed
- Plant
- Plant Derivative
- Spice
- Vegetable
- Cabbage
- Vegetable Fruit
- Gourd
- Vegetable Root
- Vegetable Stem
- Vegetable Tuber
Q2: What is ZINC Similarity Search?
ZINC is a free public resource for discovery of drug-like molecules. The database contains over twenty million commercially available molecules in biologically relevant representations that may be downloaded in popular ready-to-dock formats and subsets. The similarity search done against ZINC compares the input SMILE and each SMILE present in the database. Depending upon the similarity percentage (Eg: 90% which is the default threshold) it displays the similar compounds.
For further information please visit the ZINC database: http://zinc15.docking.org/
For further information please visit the ZINC database: http://zinc15.docking.org/
Q3: How do I use the “Flavor Pairing” app?
Refer to the Flavor Pairing section on the how to use page
Q4: What are FlavorDB Statistics?
Flavor DB statistics explain the vastness and depth of flavor DB using interactive Google Charts (https://developers.google.com/chart/).
Q5: How do I use “Visual Search”?
Refer to the Visual Search section on the how to use page
Q6: How to use “Advanced Search”?
Refer to the Advanced Search section on the how to use page
Q7: How was “The Flavor Network” constructed?
Flavor network was constructed using d3.js hierarchical edge bundeling (https://bl.ocks.org/mbostock/7607999).To address dense pattern of interrelationships due to abundance of sharing, the backbone network showing statistically significant edges is depicted.
Q8: How do you perform a similarity search?
One search option provided is to perform a molecular search by using the JSME tool. With this one can draw a molecule’s structure and search for similar molecules. For each structure that is drawn, the JSME Molecular Editor generates its corresponding SMILE value. We then run the SMILE value through Open Babel to generate a molecular fingerprint.
Then we check molecular structural similarity using Open Babel to calculate the Tanimoto Coefficient between the generated fingerprint and data from our database. We use a threshold value of 0.3 to display our results. Thus, only molecules that have a similarity level of 30% or higher are displayed.
Then we check molecular structural similarity using Open Babel to calculate the Tanimoto Coefficient between the generated fingerprint and data from our database. We use a threshold value of 0.3 to display our results. Thus, only molecules that have a similarity level of 30% or higher are displayed.
Q9: What are the different concepts used in FlavorDB and how to they relate to each other?
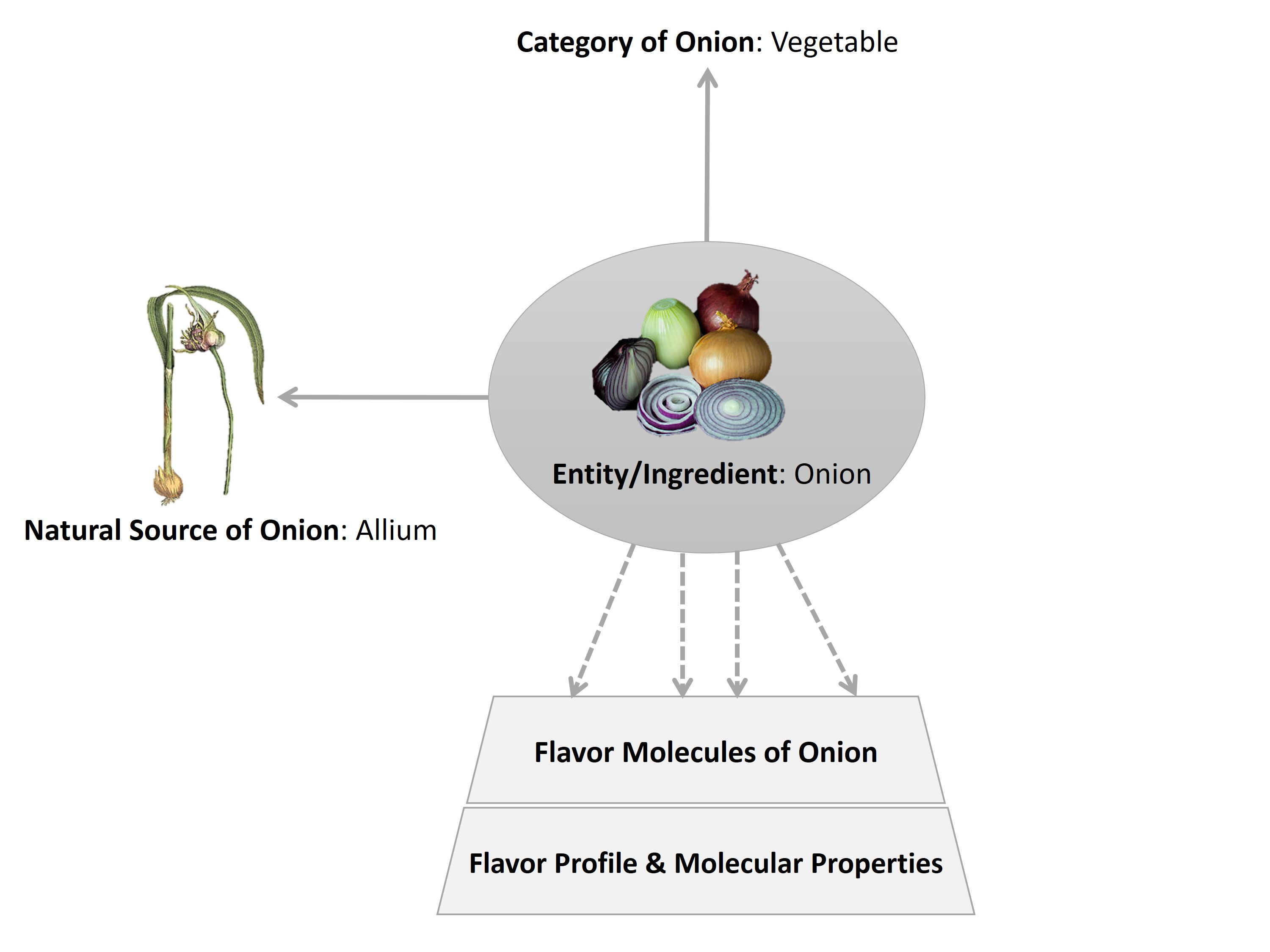
The illustration depicts relationships among different concepts incorporated in FlavorDB. The 'Entity is synonymous to an Ingredient', and is a key concept in the framework. In this example, ‘Onion’ is an Entity/Ingredient which is classified into the Category ‘Vegetable’. The natural origin (plant or animal species/genus/family/kingdom) of an ingredient is referred to as the Natural Source, which in the case of onion is ‘Allium’. Each ingredient comprises a set of flavor molecules. And further, each flavor molecule is characterized by a ‘flavor profile’ (a set of flavor terms) and by an array of ‘molecular properties’ (Physicochemical, 2D/3D and ADMET).

Q10: Prerequisites needed to run FlavorDB?
A modern web browser with JavaScript enabled.
Q11: What browsers does FlavorDB support?
FlavorDB supports all modern web browsers. But the Flavor Network can be best viewed on Chrome, Firefox and Edge
Q12: What is Tech Stack used to build FlavorDB?
Front End: HTML, CSS, JavaScript, AJAX, jQuery(v1.12.4), Bootstrap(v3.3.7), DataTables(https://datatables.net/), Google Charts(https://developers.google.com/chart/) ,D3.js
JSME Molecular Editor(http://peter-ertl.com/jsme/), Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
Back End: Python, Django, Mysql, OpenBabel(v2.4.0)
Q13: What is Jmol?
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
Jmol does not require 3D acceleration plugins. Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g. in chemistry and biochemistry. It is free and open source software, written in Java and so it runs on Windows, Mac OS X, Linux and Unix systems. Few technical advantages of Jmol are:
Jmol does not require 3D acceleration plugins. Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g. in chemistry and biochemistry. It is free and open source software, written in Java and so it runs on Windows, Mac OS X, Linux and Unix systems. Few technical advantages of Jmol are:
- Molecular 3D visualisation
- Zooming facilities
- Provision to download the viewed molecule in mol2 format
Q14: What do I need to have the Jmol files to render properly?
A modern web browser with JavaScript enabled.
Q15: What is the source of data and images?
To begin with, a list of ingredients was created using, Foodb (http://foodb.ca) and arXiv preprint arXiv:1502.03815, 2015. Each of the 951 ingredients were then manually classified into 34 categories: Additive, Animal Product, Bakery, Beverage, Beverage Alcoholic, Beverage Caffeinated, Cereal, Maize, Dairy, Dish, Essential Oil, Fish, Seafood, Flower, Fruit, Berry, Fruit Citrus, Fruit Essence, Fungus, Herb, Meat, Legume, Nut, Seed, Plant, Plant Derivative, Spice, Vegetable, Cabbage, Vegetable Fruit, Gourd, Vegetable Root, Vegetable Stem, and Vegetable Tuber. Each entity was also mapped to its natural source, with a total of 532 unique sources being identified. The details of entities and their natural sources, related images, and scientific classification were obtained from Wikipedia using Python’s BeautifulSoup4 library (https://www.crummy.com/software/BeautifulSoup) and MediaWiki’s action API (MediaWiki The Free Wiki Engine).
The data of flavor molecules for each of these ingredients were compiled via literature survey, flavor resources, and previously reported data:
The chemical identifiers of molecules were obtained from various sources, and were standardized to procure their CAS (Chemical Abstract Service) numbers. CAS numbers were then mapped to their corresponding PubChem IDs, as the former are often degenerate with multiple CAS numbers pointing to the same molecule, and some pointing to multiple molecules. Thus, PubChem ID was used as the unique primary key for every flavor molecule. Using the PubChem ID, compound identifiers (such as common name, IUPAC, Canonical SMILES), physicochemical properties and 2D images were obtained from PubChem REST API (https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html). The flavor profile of the molecule (FEMA Flavor Profile, FEMA Number, Taste, and Odor) was downloaded from PubChem.
Further 2D/3D, ADMET and physicochemical properties as well as Mol2 files for all 25595 molecules were obtained using Discovery Studio 4.0. The functional groups were obtained using Checkmol software. Functional group refers to an atom, or a group of atoms that have similar chemical properties whenever they occur in different compounds. Thus, it defines the characteristic physical and chemical properties of families of organic compounds.
The data of flavor molecules for each of these ingredients were compiled via literature survey, flavor resources, and previously reported data:
- Burdock,G.A. (2010) Fenaroli’s handbook of flavor ingredients.
- Ahn,Y.-Y., Ahnert,S.E., Bagrow,J.P. and Barabási,A.-L. (2011) Flavor network and the principles of food pairing. Sci. Rep., 1, 196.
- Jain,A., Rakhi,N.K. and Bagler,G. (2015) Analysis of food pairing in regional cuisines of India. PLoS One, 10.
- Jain,A., Rakhi,N.K. and Bagler,G. (2015) Spices form the basis of food pairing in Indian cuisine. arXiv:1502.03815.
- www.foodb.ca
- Arn,H. and Acree,T.E. (1998) Flavornet: A database of aroma compounds based on odor potency in natural products. Dev. Food Sci., 40, 27.
- Ahmed,J., Preissner,S., Dunkel,M., Worth,C.L., Eckert,A. and Preissner,R. (2011) SuperSweet-A resource on natural and artificial sweetening agents. Nucleic Acids Res., 39.
- Wiener,A., Shudler,M., Levit,A. and Niv,M.Y. (2012) BitterDB: A database of bitter compounds. Nucleic Acids Res., 40, 413–419, D377-82.
The chemical identifiers of molecules were obtained from various sources, and were standardized to procure their CAS (Chemical Abstract Service) numbers. CAS numbers were then mapped to their corresponding PubChem IDs, as the former are often degenerate with multiple CAS numbers pointing to the same molecule, and some pointing to multiple molecules. Thus, PubChem ID was used as the unique primary key for every flavor molecule. Using the PubChem ID, compound identifiers (such as common name, IUPAC, Canonical SMILES), physicochemical properties and 2D images were obtained from PubChem REST API (https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html). The flavor profile of the molecule (FEMA Flavor Profile, FEMA Number, Taste, and Odor) was downloaded from PubChem.
Further 2D/3D, ADMET and physicochemical properties as well as Mol2 files for all 25595 molecules were obtained using Discovery Studio 4.0. The functional groups were obtained using Checkmol software. Functional group refers to an atom, or a group of atoms that have similar chemical properties whenever they occur in different compounds. Thus, it defines the characteristic physical and chemical properties of families of organic compounds.
Q16: What are Functional Groups and how were they obtained?
Functional Groups:- The functional group is an atom, or a group of atoms that has similar chemical properties whenever it occurs in different compounds. It defines the characteristic physical and chemical properties of families of organic compounds. (http://goldbook.iupac.org/F02555.html)
Checkmol: - A free and an open source tool, checkmol, detect and assign the functional group information on any small molecules with 2D coordinates. The checkmol is a command-line utility program, which reads molecular structure files in different formats and analyzes the input molecule for the presence of various functional groups. The output text can be easily placed into a database table, permitting the creation of chemical databases with a functional group search option.
Checkmol: - A free and an open source tool, checkmol, detect and assign the functional group information on any small molecules with 2D coordinates. The checkmol is a command-line utility program, which reads molecular structure files in different formats and analyzes the input molecule for the presence of various functional groups. The output text can be easily placed into a database table, permitting the creation of chemical databases with a functional group search option.
Analysis of Functional Groups in Organic Molecules, http://merian.pch.univie.ac.at/~nhaider/cheminf/cmmm.html
-
N. Haider, Functionality pattern matching as an efficient complementary structure/reaction search tool: an open-source approach, Molecules 15 (8) (2010) 5079–5092.( http://www.mdpi.com/1420-3049/15/8/5079)
Q17: Do you use cookies?
We are using cookies to provide statistics that help us give best experience for our site.
Q18: Can we use FlavorDB in mobiles?
Yes, just visit flavorDB from your mobile browser. But the data visualizations such as flavor network are best viewed on desktop.
Q19: How do I access the data from FlavorDB?
Data is accessible for molecules as well as entities/ingredients. These can be downloaded in JSON format from the ‘Molecular & Flavor Profile’ tab (More Info.) and the entity page respectively. The molecules’ data can also be obtained in Mol2, 2D Image and SDF formats. These data are available under a Creative Commons License Attribution-NonCommercial-ShareAlike 3.0 Unported (CC BY-NC-SA 3.0).


Q20: How do I contribute to data?
You may contact us at bagler+FlavorDB@iiitd.ac.in for errata or suggesting information of flavor profile and ingredient association with relevant references.
You may also fill the following forms to report errors or giving suggestions.
Suggestions Form for reporting new flavor molecules or Entity/Ingredient associations:
https://docs.google.com/forms/d/e/1FAIpQLSe9m_rNojup_dQohU3gHA-ByYbbbB9-x6bv4knyxu0ZQ2ClTA/viewform
Errata Form to report errors in data:
https://docs.google.com/forms/d/e/1FAIpQLSfRYt6D4-c9UKGFpvvucqRvV871pMix6mnC1cskT9CXcEiLLw/viewform
You may also fill the following forms to report errors or giving suggestions.
Suggestions Form for reporting new flavor molecules or Entity/Ingredient associations:
https://docs.google.com/forms/d/e/1FAIpQLSe9m_rNojup_dQohU3gHA-ByYbbbB9-x6bv4knyxu0ZQ2ClTA/viewform
Errata Form to report errors in data:
https://docs.google.com/forms/d/e/1FAIpQLSfRYt6D4-c9UKGFpvvucqRvV871pMix6mnC1cskT9CXcEiLLw/viewform
Q21: Under what license is FlavorDB available?

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
Q21: How to Cite us?
Neelansh Garg†, Apuroop Sethupathy†, Rudraksh Tuwani†, Rakhi NK†, Shubham Dokania†, Arvind Iyer†, Ayushi Gupta†, Shubhra Agrawal†, Navjot Singh†, Shubham Shukla†, Kriti Kathuria†, Rahul Badhwar, Rakesh Kanji, Anupam Jain, Avneet Kaur, Rashmi Nagpal, and Ganesh Bagler*, FlavorDB: A database of flavor molecules, Nucleic Acids Research, gkx957, (2017). †Equal contribution *Corresponding Author
Q22: Who is the team behind FlavorDB?
| Name | Position | Affiliation | Contribution |
|---|---|---|---|
| Ganesh Bagler | Project Head | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Idea conception, Project design and management, Database design and implementation |
| Neelansh Garg | Summer Research Intern | USICT, Guru Gobind Singh Indraprastha University, New Delhi | Database design, Development of FlavorDB Web Resource, Data Mining, Data Visualisation and Data Analytics |
| Apuroop Sethupathy | Summer Research Intern | Ashoka University, Sonepat | Database design, Development of FlavorDB Web Resource, Data Mining, Data Visualisation and Data Analytics |
| Rudraksh Tuwani | Summer Research Intern | Sri Venkateswara College, Delhi University | Natural Language Processing, feature extraction, Data Mining and Data Analytics |
| Rakhi N K | PhD Research Scholar | Department of Bioscience and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur | Manual data compilation, and curation, Quality Check, food pairing analysis |
| Shubham Dokania | Summer Research Intern | Delhi Technological University, New Delhi | Feature extraction and data mining |
| Arvind Iyer | M.Tech (Computational Biology) Student | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Database design, Development of FlavorDB Web Resource, Data Mining |
| Ayushi Gupta | M.Tech (Computational Biology) Student | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Data compilation, curation and Data structure design |
| Shubhra Agrawal | M.Tech Student | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Data compilation, curation and Data structure design |
| Navjot Singh | Summer Research Intern | Delhi Technological University, New Delhi | Food pairing analysis |
| Shubham Shukla | Summer Research Intern | Northern India Engineering College, Guru Gobind Singh Indraprastha University, New Delhi | Data extraction and Quality Check |
| Kriti Kathuria | Summer Research Intern | Maharaja Agrasen College, Delhi University, New Delhi | Data retrieval and compilation, Quality Check |
| Anupam Jain | M.Tech. (Systems Science) Student | Department of Bioscience and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur | Data compilation and food pairing analysis |
| Rahul Badhwar | PhD Research Scholar | Department of Bioscience and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur | Molecular properties extraction |
| Rakesh Kanji | PhD Research Scholar | Department of Bioscience and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur | Molecular properties extraction |
| Avneet Kaur | Research Intern | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Data compilation assistance |
| Rashmi Nagpal | Research Intern | Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT-Delhi), New Delhi | Data compilation assistance |