SpiceRx integrates evidence-based knowledge pertaining to the health impacts of culinary spices and herbs and their phytochemicals. It provides a systematic compilation of tripartite relationships between culinary spices/herbs, their phytochemicals and diseases.
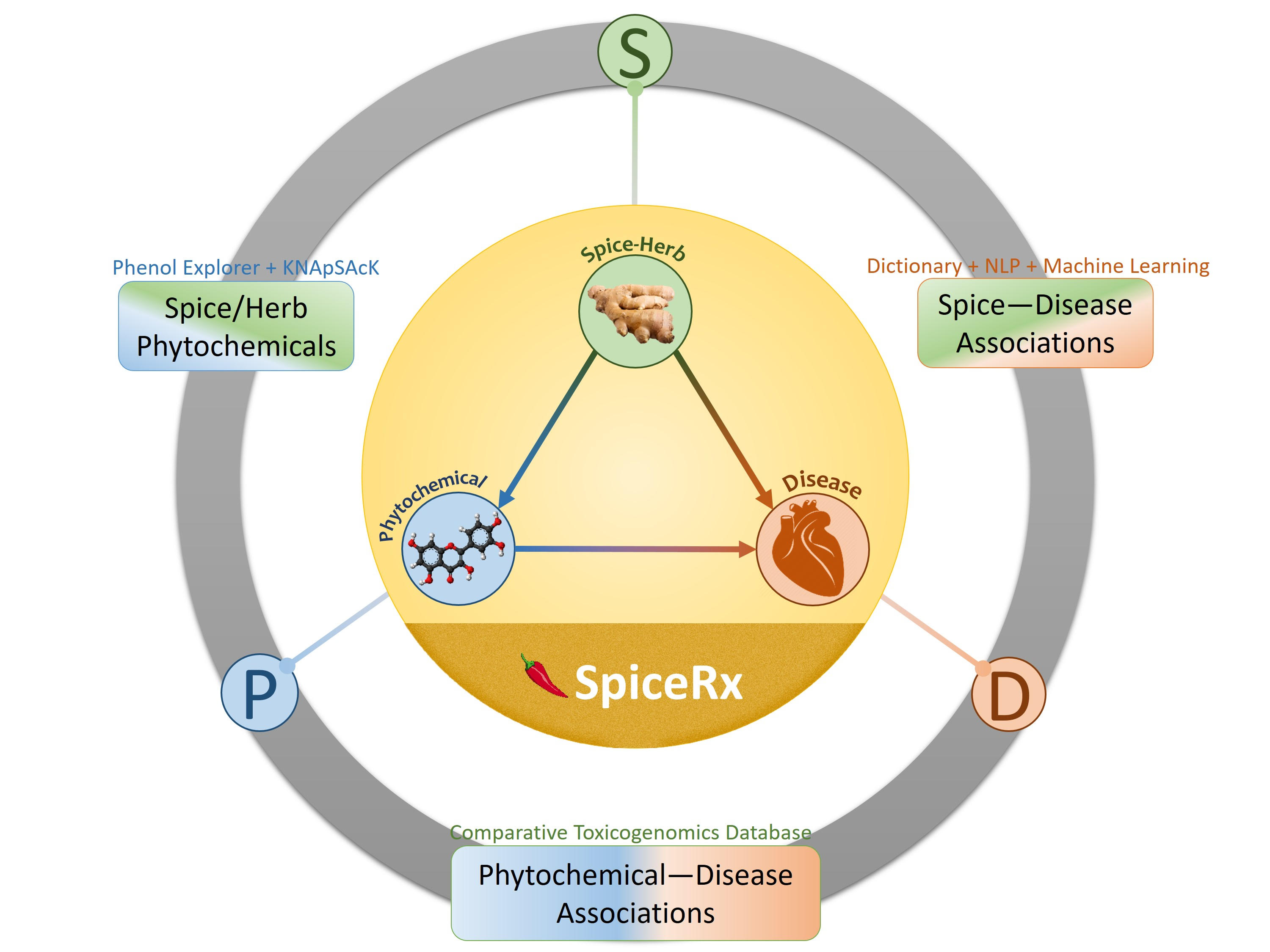
SpiceRx facilitates three types of queries based on spices/herbs, diseases and phytochemicals. Given one, it displays relationships with the remaining two.
SpiceRx data was compiled starting with a total of 188 culinary spices and herbs used in recipes worldwide. Out of these, 152 have documented disease associations.
Data for 866 phytochemicals from 142 culinary spices/herbs were compiled. Among these, 570 phytochemicals are bioactive, involved in 2042 spice-phytochemical associations.
Spice-disease associations were text mined from 23 million MEDLINE abstracts indexed in PubMed till July 2017.
A dictionary was compiled from various sources such as FooDB, Wikipedia, PFAF, FPI, and FlavorDB.
MeSH is a controlled vocabulary developed by the National Library of Medicine. It classifies diseases into hierarchical categories, subcategories, and diseases.
SpiceRx includes 188 culinary spices and herbs, of which 152 have disease associations as documented in the literature.
SpiceRx supports all modern web browsers.
SpiceRx is implemented with Django and PostgreSQL. The frontend uses HTML, CSS, JavaScript, AJAX, jQuery, JSME Molecular Editor, Bootstrap, Jmol, DataTables, and Google Charts. An Apache server routes requests and enables compression.
A modern web browser with JavaScript enabled.
We use cookies to collect statistics that help us improve your experience.
Jmol is an open-source Java viewer for chemical structures in 3D. It does not require 3D acceleration plugins and is used for both teaching and research in chemistry and biochemistry.
A modern web browser with JavaScript enabled.
All material on this website is research-based and provided for information only. It should not be construed as medical advice—consult appropriate health professionals for any concerns.
| Name | Position | Affiliation | Contribution |
|---|---|---|---|
| Ganesh Bagler | Project Head | Center for Computational Biology, IIIT-Delhi, New Delhi | Idea conception, project design and management, database design and implementation |
| Rakhi N K | PhD Research Scholar | Department of Bioscience and Bioengineering, IIT Jodhpur | Manual data compilation, annotation and curation, quality check, analysis |
| Rudraksh Tuwani | Research Assistant | Center for Computational Biology, IIIT-Delhi | Text mining, database design, web resource development, data visualization and analytics |
| Neelansh Garg | Summer Research Intern | USICT, Guru Gobind Singh Indraprastha University, New Delhi | Database design, web resource development, data visualization and analytics |
| Jagriti Mukherjee | M.Tech student | Center for Computational Biology, IIIT-Delhi, New Delhi | Annotations |
| Kshitija Randive | M.Tech student | Computer Science and Engineering, IIIT-Delhi, New Delhi | Database redesign, Data visualization and Analytics |
| Abhishek Gond | M.Tech student | Computer Science and Engineering, IIIT-Delhi, New Delhi | Frontend, UI Upgradation and redesign |